
Photo Adapted from: Tetra Images/Alamy
Before ancient DNA exposed the sexual proclivities of Neanderthals or the ancestry of the first Americans, there was the quagga.
An equine oddity with the head of a zebra and the rump of a donkey, the last quagga (Equus quagga quagga) died in 1883. A century later, researchers published1
around 200 nucleotides sequenced from a 140-year-old piece of quagga
muscle. Those scraps of DNA — the first genetic secrets pulled from a
long-dead organism — revealed that the quagga was distinct from the
mountain zebra (Equus zebra).
More
significantly, the research showed that from then on, examining fossils
would no longer be the only way to probe extinct life. “If the long-term
survival of DNA proves to be a general phenomenon,” geneticists Russell
Higuchi and Allan Wilson of the University of California, Berkeley, and
their colleagues noted in their quagga paper
1, “several fields including palaeontology, evolutionary biology, archaeology and forensic science may benefit.”
At
first, progress was fitful. Concerns over the authenticity of
ancient-DNA research fuelled schisms in the field and deep scepticism
outside it. But this has faded, thanks to laboratory rigour that borders
on paranoia and sequencing techniques that help researchers to identify
and exclude contaminating modern DNA.
These
advances have fostered an ancient-genomics boom. In the past year,
researchers have unveiled the two oldest genomes on record: those of a
horse that had been buried in Canadian permafrost for around 700,000
years2, and of a roughly 400,000-year-old human relative from a Spanish cavern3. A Neanderthal sequence every bit as complete and accurate as a contemporary human genome has been released4, as has the genome of a Siberian child connecting Native Americans to Europeans5.
Enabling
this rush are technological improvements in isolating, sequencing and
interpreting the time-ravaged DNA strands in ancient remains such as
bones, teeth and hair. Pioneers are obtaining DNA from ever older and
more degraded remains, and gleaning insight about long-dead humans and
other creatures. And now ancient DNA is set to move from the clean-rooms
of specialists to the labs of archaeologists, population geneticists
and others. Thirty years after the quagga led the way,
Nature looks to the field's future.
A million-year-old genome
Ludovic
Orlando, an evolutionary biologist at the University of Copenhagen, had
low expectations when he started sequencing DNA from a
560,000-to-780,000-year-old horse leg bone. His colleague, Eske
Willerslev, had discovered the bone buried in the permafrost of the
Canadian Yukon in 2003. Then he had chucked it into a freezer, waiting
for technological improvements that would allow the bone's degraded DNA
to be read. (Freezers in ancient-DNA labs brim with such 'wait and see'
samples.)
On a Sunday evening in 2010,
Willerslev called Orlando to say that the time had come. Orlando was
unconvinced: “I started the project with the firm intention of proving
that it was not possible,” he says.
Sequencing
ancient DNA is a battle against time. After an organism dies, the long
strands of its DNA fissure into ever shorter pieces, helped along by
DNA-munching enzymes. Low temperatures slow this process, but eventually
the strands become so short that they contain little information.
To
read the horse's genome, Orlando needed to shepherd useful DNA
fragments through the harsh enzymatic treatments used to extract them
and ready them for sequencing. Orlando and his team found that the
preparation lost vast quantities of fragments. But with a few tweaks to
the experimental protocol, such as reducing the extraction temperature,
the researchers captured ten times more scraps of DNA than before — and
produced a draft of the oldest genome on record
2.
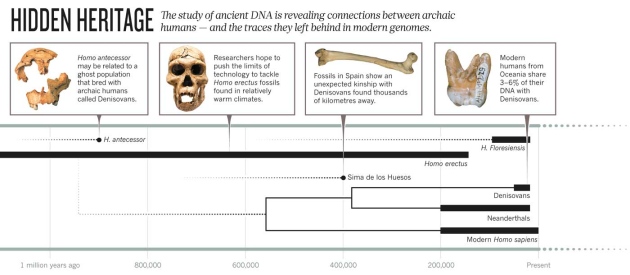
Photos (L–R): Javier Trueba/MSF/SPL; Markus Schieder/Alamy; Ref. 3; D. Reich et al. Nature 468, 1053–1060 (2010)
Using a similar approach, Svante Pääbo, a
geneticist at the Max Planck Institute for Evolutionary Anthropology in
Leipzig, Germany, and his team turned their attention to
400,000-year-old remains from the Sima de los Huesos cavern in northern
Spain, which may have been a burial pit for recent relatives of modern
humans called hominins (see
'Hidden heritage').
In the pit, the bones remained at stable, low temperatures, slowing the
breakdown of DNA. “If you could have told the hominins where to leave
their bones, you may have chosen that site,” says Matthias Meyer, a
molecular biologist at Pääbo's institute who is leading the efforts.
Last December, the team reported
3
roughly 16,300 letters of a Sima de los Huesos individual's
mitochondrial genome — the DNA from power-generating structures in its
cells. The sequence revealed an unexpected relationship between the Sima
de los Huesos remains and the Denisovans, an archaic group of humans
that Pääbo's team had discovered in Russia's Altai Mountains thousands
of kilometres away. Meyer and his colleagues hope to improve their
methods enough to obtain some or all of the Sima de los Huesos
individual's nuclear genome, the DNA from the nuclei of its cells. “It
must be possible,” says Meyer. “I won't rest until this has been done.”
It
is now a matter of when, not if, someone will produce a genome from an
Arctic animal buried in permafrost for longer than 1 million years, says
Meyer. But he and Pääbo want to push the limits of ancient DNA in
hominin specimens from warmer locales, such as fossils of
Homo erectus,
the common ancestor of humans and Neanderthals, found in Asia. And
Orlando says that researchers may have luck using new extraction
techniques on previously vexing remains such as Egyptian mummies or
Homo floresiensis,
a small hominin at least 18,000 years old that was found in a cave on
the Indonesian island of Flores. “It opens a great number of places
where there are lots of important stories going on, such as the Middle
East or the tropics,” he says.
Ghosts in the code
A
few years ago, David Reich discovered a ghost. Reich, a population
geneticist at Harvard Medical School in Boston, Massachusetts, and his
team were reconstructing the history of Europe using genomes from modern
people, when they found a connection between northern Europeans and
Native Americans. They proposed that a now-extinct population in
northern Eurasia had interbred with both the ancestors of Europeans and a
Siberian group that later migrated to the Americas
6.
Reich calls such groups ghost populations, because they are identified
by the echoes that they leave in genomes — not by bones or ancient DNA.
Ghost
populations are the product of statistical models, and as such should
be handled with care when genetic data from fossils are lacking, says
Carlos Bustamante, a population geneticist at Stanford University in
California. “When are we reifying something that's a statistical
artefact, versus when are we understanding something that's a true
biological event?”
Sometimes these statistical spectres get a body. Last year, Willerslev's team reported
5
the genome from 24,000-year-old remains dubbed the Mal'ta boy. The
results showed that the boy, who had been found in central Siberia, came
from a population related to both modern Native Americans and modern
Europeans, matching Reich's prediction (see
Nature http://doi.org/r2b; 2013). “It's a spectacular find,” he says.
Ghost
populations also lurk in ancient DNA. While analysing high-quality
genomes of a Neanderthal and a Denisovan, a team led by Reich and
Montgomery Slatkin at the University of California, Berkeley, noticed a
peculiar pattern: present-day sub-Saharan Africans are more closely
related to Neanderthals than they are to Denisovans
4.
But evidence from other ancient genomes suggested that the two archaic
groups were equally related to present-day Africans. After weighing the
possibilities, the scientists realized that they might have uncovered
another ghost population.
The puzzle could be
solved, they theorized, if Denisovans had interbred with a species that
had left Africa perhaps more than 1 million years ago and branched off
from the common ancestor of humans, Neanderthals and Denisovans.
Subsequent Denisovans would have inherited DNA sequences that
present-day Africans lack, explaining why Neanderthals seem to be closer
kin to Africans.
Reich's team is analysing
genetic signatures in humans with Denisovan DNA to establish when the
Denisovans mated with this mystery population — information that could
narrow the range of fossils to which it might belong. Genomes studied by
Pääbo's lab, principally the Sima de los Huesos remains, may also
reveal clues.
Reich is not the only one
conjuring ghosts. Chris Stringer, a palaeoanthropologist at the Natural
History Museum in London, has proposed that the 900,000-year-old hominin
Homo antecessor, known from fossils found near Sima de los
Huesos, could be part of the ghost population. If it had interbred with
an ancestor of the Denisovans and the Sima de los Huesos hominins, it
could explain the relationship between the two groups of remains.
Testing that hypothesis would require the elusive Sima de los Huesos
nuclear DNA. But Reich is optimistic that Pääbo and his team will pull
it off. “They've done miracles before in that lab and they may succeed
again.”
The Neanderthal within
“We
don't need bones necessarily to find ancient DNA,” says Josh Akey, a
population geneticist at the University of Washington in Seattle. “We
can find the remnants of ancient DNA floating around in contemporary
populations.”
If early human populations bred
with Neanderthals and Denisovans, their descendants should carry short
segments of archaic-human DNA. Researchers such as Akey are beginning to
catalogue these segments to learn about the biology of archaic humans.
Unlike the hunt for ghost populations, which relies on statistical
population models, this approach allows researchers to identify specific
regions of the genome acquired by interbreeding.
“With tools that make sequencing ancient DNA cheaper and easier, the field is becoming more egalitarian.”
In January, independent teams led by Akey
7 and Reich
8
pieced together a substantial portion — about 20% and 40% respectively —
of the Neanderthal genome from bits lurking in the genomes of hundreds
of living humans. Their research indicated that some Europeans and
Asians had gained genes involved in skin and hair from Neanderthals,
possibly helping their ancestors to adapt to cold climates by providing
thicker skin, more hair and fewer pores (see
Nature http://doi.org/rz9;
2014). But giant swathes of the modern genomes were devoid of
Neanderthal ancestry, hinting that many Neanderthal genes might have
been harmful in modern humans. Akey's team identified
7 one such region around the gene
FOXP2,
which is involved in speech and language. “It's extremely compelling
evidence that there were fitness costs to interbreeding,” he says.
These
discoveries are only the beginning. The Akey and Reich teams found that
the genomes of east Asians possess, on average, slightly more
Neanderthal DNA than do people of European ancestry. Akey sees this as
possible evidence that Neanderthals interbred with ancient humans on at
least two separate occasions: once with the ancestors of all Eurasians,
and later with a population ancestral only to east Asians. And Akey
believes that humans are likely to bear genetic scraps from other
extinct species, including some that interbred with the ancestors of
humans in sub-Saharan Africa.
Ancient DNA for the masses
For
much of the past 30 years, the sensitivity of the polymerase chain
reaction (PCR), the method used to amplify ancient DNA, made it prone to
contamination. The field's leaders often greeted the work of outsiders
with suspicion, earning some of them the title 'the PCR police'. And in
recent years, palaeogenomics has been the domain of specialist labs such
as Pääbo's, with the expertise and money to obtain and screen hundreds
of fossils to find the few that yield enough DNA to sequence an entire
genome.
That is set to change. New procedures
mean that researchers can now reliably obtain DNA from all but the most
degraded samples, and then sequence only the portions of a genome that
they are interested in. “I'm still surprised that there are so few labs
in the world that do this,” says Johannes Krause, a palaeogeneticist at
the University of Tübingen, Germany, who led much of the Denisovan work
while in Pääbo's lab. “It's not rocket science.”
Gradually,
new researchers are entering the field. “If I can break in, then anyone
can,” jokes Bustamante. His research originally focused on ancestry in
current human populations. Then, a few years ago, he got a phone call
about a mummy.
An international team had
sequenced the genome of Ötzi, a 5,300-year-old frozen corpse found in
the Tyrolean Alps of Italy in 1991. The researchers wondered if
Bustamante could help them to make sense of the ice-man's ancestry.
Together, they showed that Ötzi was more closely related to humans who
now live in Sardinia and Corsica than those in central Europe, evidence
that the population of Europe when he was alive looked very different to
how it does today9.
Bustamante
has since plunged into the world of ancient DNA. His team is sequencing
samples that chart the arrival of farming in Bulgaria, the
transatlantic slave trade in the Americas and dog domestication. The
group is developing tools to make sequencing ancient DNA cheaper and
easier. “We want to democratize the field,” says Bustamante.
Reich,
too, sees ancient DNA becoming more egalitarian. His lab's growing
interest in areas of human history such as the advent of agriculture or
the history of the Indian subcontinent has led it to analyse — often in
bulk — remains less rarefied than the scarce Neanderthal samples that
first lured him to the field.
Last year, Reich was part of a team that reported
10
an analysis of mitochondrial DNA from 364 European samples between
5,500 and 1,550 years old, to identify major population shifts in
Neolithic Europe. Ancient genomics is also set to solve long-standing
questions about when and where humans domesticated animals such as dogs,
cattle and chickens. A 2013 study
11
of 18 mitochondrial genomes from ancient dogs and wolves, for instance,
suggested that European hunter-gatherers domesticated wolves from a
population that is now extinct.
Researchers
are also returning to the questions that launched the field 30 years
ago. Around the time that Orlando's team began sequencing the
700,000-year-old horse, it also turned its attention to a much younger
sample — from the quagga.
The effort to
sequence the full quagga genome is part of large project to understand
the evolutionary relationship between living and extinct horses, zebras
and donkeys, and to identify the genetic basis for certain traits. “I
was thinking it would be cool to do the oldest, but also the first —
where ancient DNA started,” says Orlando. “It shows the progress the
field has made.”


