New human gene tally reignites debate
Nova contagem de genes humanos reacende debate
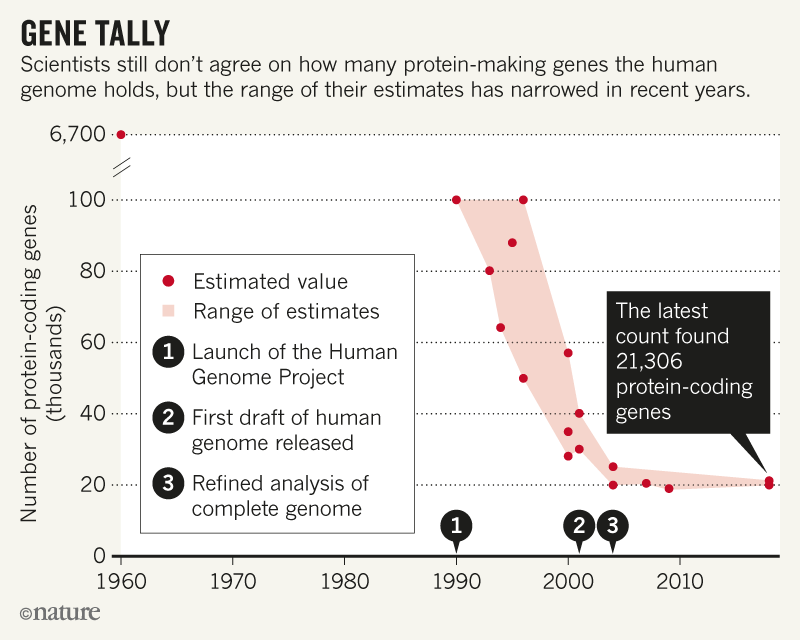
That was in 2000, when a draft human genome sequence was still in the works; geneticists were running a sweepstake on how many genes humans have, and wagers ranged from tens of thousands to hundreds of thousands. Almost two decades later, scientists armed with real data still can’t agree on the number — a knowledge gap that they say hampers efforts to spot disease-related mutations.
The latest attempt to plug that gap uses data from hundreds of human tissue samples and was posted on the BioRxiv preprint server on 29 May1. It includes almost 5,000 genes that haven’t previously been spotted — among them nearly 1,200 that carry instructions for making proteins. And the overall tally of more than 21,000 protein-coding genes is a substantial jump from previous estimates, which put the figure at around 20,000.
But many geneticists aren’t yet convinced that all the newly proposed genes will stand up to close scrutiny. Their criticisms underscore just how difficult it is to identify new genes, or even define what a gene is.
“People have been working hard at this for 20 years, and we still don’t have the answer,” says Steven Salzberg, a computational biologist at Johns Hopkins University in Baltimore, Maryland, whose team produced the latest count.
Hard to pin down
In 2000, with the genomics community abuzz over the question of how many human genes would be found, Ewan Birney launched the GeneSweep contest. Birney, now co-director of the European Bioinformatics Institute (EBI) in Hinxton, UK, took the first bets at a bar during an annual genetics meeting, and the contest eventually attracted more than 1,000 entries and a US$3,000 jackpot. Bets on the number of genes ranged from more than 312,000 to just under 26,000, with an average of around 40,000. These days, the span of estimates has shrunk — with most now between 19,000 and 22,000 — but there is still disagreement (See 'Gene Tally').
Salzberg’s team used data from the Genotype-Tissue Expression (GTEx) project, which sequenced RNA from more than 30 different tissues taken from several hundred cadavers. RNA is the intermediary between DNA and proteins. The researchers wanted to identify genes that encode a protein and those that don’t but still serve an important role in cells. So they assembled GTEx’s 900 billion tiny RNA snippets and aligned them with the human genome.
Just because a stretch of DNA is expressed as RNA, however, does not necessarily mean it’s a gene. So the team attempted to filter out noise using a variety of criteria. For example, they compared their results with genomes from other species, reasoning that sequences shared by distantly related creatures have probably been preserved by evolution because they serve a useful purpose, and so are likely to be genes.
The team was left with 21,306 protein-coding genes and 21,856 non-coding genes — many more than are included in the two most widely used human-gene databases. The GENCODE gene set, maintained by the EBI, includes 19,901 protein-coding genes and 15,779 non-coding genes. RefSeq, a database run by the US National Center for Biotechnology Information (NCBI), lists 20,203 protein-coding genes and 17,871 non-coding genes.
Kim Pruitt, a genome researcher at the NCBI in Bethesda, Maryland, and a former head of RefSeq, says the difference is probably due in part to the volume of data that Salzberg’s team analysed. And there’s another major difference. Both GENCODE and RefSeq rely on manual curation — a person reviews the evidence for each gene and makes a final determination. Salzberg’s group relied solely on computer programmes to sift the data.
“If people like our gene list, then maybe a couple years from now we’ll be the arbiter of human genes,” says Salzberg.
Tricky tally
But many scientists say they need more evidence to be convinced that the list is accurate. Adam Frankish, a computational biologist at the EBI who coordinates the manual annotation of GENCODE, says that he and his group have scanned about 100 of the protein-coding genes identified by Salzberg’s team. By their assessment, only one of those seems to be a true protein-coding gene.
And Pruitt’s team looked at about a dozen of the Salzberg group’s new protein-coding genes, but didn’t find any that would meet RefSeq’s criteria. Some overlapped with regions of the genome that seem to belong to retroviruses that invaded our ancestors’ genomes; others belong to other repetitive stretches, which are rarely translated into proteins.
But Salzberg says that some repetitive sequences can be considered genes. One example is ERV3-1, which appears in RefSeq and encodes a protein that is overexpressed in colorectal cancer. Salzberg also acknowledges that the new genes on his team’s list will require validation by his team and others.
Further confounding counting efforts is the imprecise and changing definition of a gene. Biologists used to see genes as sequences that code for proteins, but then it became clear that some non-coding RNA molecules have important roles in cells. Judging which are important — and should be deemed genes — is controversial, and could explain some of the discrepancies between Salzberg’s count and others.
Still, it’s likely that at least some of the genes identified by Salzberg’s group will turn out to be valid, says Emmanouil Dermitzakis, a geneticist at the University of Geneva in Switzerland, who co-chairs the GTEx project. He isn’t surprised that the team’s count for protein-coding genes is a 5% increase on previous tallies, given the gargantuan size of the GTEx data set.
Having an accurate tally of all human genes is important for efforts to uncover links between genes and disease. Uncounted genes are often ignored, even if they contain a disease-causing mutation, Salzberg says. But hastily adding genes to the master list can pose risks, too, says Frankish. A gene that turns out to be incorrect can divert geneticists’ attention away from the real problem.
Still, the inconsistencies in the number of genes from database to database are problematic for researchers, Pruitt says. “People want one answer,” she adds, “but biology is complex.”
Nature 558, 354-355 (2018)
doi: 10.1038/d41586-018-05462-w

Nenhum comentário:
Postar um comentário
Observação: somente um membro deste blog pode postar um comentário.