Reassessing evidence of life in 3,700-million-year-old rocks of Greenland
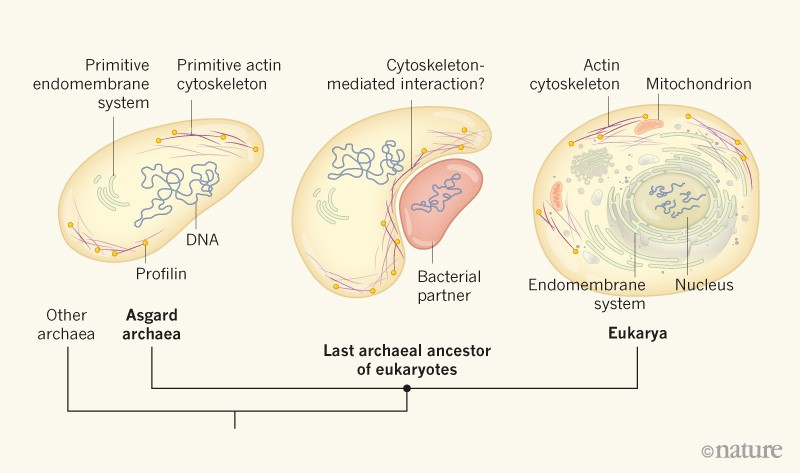
Eukaryotic cells, which carry their DNA in a nucleus, are thought to have evolved from a merger between two other organisms — an archaeal host cell1–3 and a bacterium from which eukaryotic organelles called mitochondria emerged4. Some insights into the biological properties of the host have come from the closest known archaeal relatives of eukaryotes, the Asgard superphylum5,6. The genomes of organisms belonging to this archaeal group encode a suite of proteins typically involved in functions or processes thought to be eukaryote-specific. The functions of these ‘eukaryotic genes’ in Asgard archaea have been elusive, but in a paper in Nature, Akıl and Robinson7 provide evidence that some of them encode proteins that are structurally and functionally similar to their eukaryotic counterparts.
Apart from their nucleus and energy-producing mitochondria, eukaryotic cells are characterized by a complex internal system of membrane-bound compartments (the endomembrane system), and by a dynamic network of proteins such as actin, called the cytoskeleton. The latter gives the cells their shape and structure, but is also involved in a variety of cellular processes specific to eukaryotes8. These features are thought to have been present in the last common ancestor of all eukaryotes, which lived about 1.8 billion years ago9, but no life forms have been found that represent an intermediate between eukaryotes and their bacterial and archaeal ancestors. The seemingly sudden emergence of cellular complexity in the eukaryotic lineage is a conundrum for evolutionary biologists.
Several of the proteins produced by Asgard archaea are evolutionarily related to proteins that in eukaryotes modulate complex cellular processes5,6. The identification of these proteins raised the question of whether Asgard archaea have some primitive versions of certain eukaryotic properties. If they do, it would suggest that the last archaeal ancestor of eukaryotes already displayed a certain — albeit probably limited — degree of cellular complexity reminiscent of eukaryotes.
Experiments to support such ideas are complicated by the fact that evidence for the existence of the four known Asgard lineages (Lokiarchaeota, Odinarchaeota, Thorarchaeota and Heimdallarchaeota)5,6 is based solely on metagenomics analyses. The cells have yet to be observed under a microscope, and have not been cultured in vitro. Nevertheless, Akıl and Robinson were determined to gain insight into the properties of Asgard proteins related to the eukaryotic proteins actin and profilin. In eukaryotes, profilin regulates the polymerization of actin into filaments of the cytoskeleton. These filaments have pivotal roles in processes that include vesicle and organelle movement, cell-shape formation and phagocytosis8, in which cells ingest foreign particles or other cells.
To produce Asgard profilins, Akıl and Robinson expressed these proteins in the bacterium Escherichia coli using a circular DNA molecule called a plasmid that harboured the profilin-encoding genes. They then purified the proteins and studied their structures using X-ray crystallography. Asgard profilins share limited amino-acid sequence identity with their eukaryotic counterparts. Nonetheless, the authors found that the structure of lokiarchaeal profilin is topologically similar to that of human profilin, although some structural divergences could be observed. This confirms that Asgard and eukaryotic profilins are indeed evolutionarily related, albeit distantly.
Next, the researchers set out to investigate whether Asgard profilins could interact with Asgard actins. Unfortunately, despite considerable efforts, they were unable to produce functional Asgard actin. As an alternative, they therefore carried out in vitro and co-crystallization experiments to test whether Asgard profilins could interact with eukaryotic actins. Remarkably, despite being separated by 2 billion to 3 billion years of evolution9, several of the Asgard profilins bound to mammalian actin and regulated its polymerization kinetics. Asgard and mammalian profilins seem to have similar effects on mammalian actin, although the Asgard proteins act less efficiently. These results suggest that Asgard archaea harbour a profilin-regulated actin cytoskeleton — a cellular feature generally regarded as a defining characteristic of eukaryotic cells (Fig. 1).
The inference of a primitive dynamic actin cytoskeleton in Asgard archaea sheds light on the biological properties of the ancestor of eukaryotes. In eukaryotic cells, the energy required to dynamically regulate actin is mainly provided by mitochondria10. Although the energetic and metabolic properties of Asgard archaea are currently unknown, they certainly lack the firepower that mitochondria provide. A profilin-regulated actin cytoskeleton in the archaeal ancestor of eukaryotes is therefore unlikely to sustain energy-consuming processes such as phagocytosis.
But was the energy provided by mitochondria necessarily the ultimate driving force for the emergence of complex cellular features in eukaryotes? Archaea such as Ignicoccus hospitalis, along with several types of bacterium, have independently evolved endomembrane systems11. Because these lineages lack mitochondria, energetic constraints can be ruled out as a limiting factor in the emergence of such a system. It is therefore feasible that Asgard archaeal cells produce sufficient energy to harbour both a primitive endomembrane system and undergo actin-driven membrane and cell-shape deformation. Perhaps the latter ability could have facilitated the symbiotic interaction between the Asgard-related host cell and the bacterial ancestor of mitochondria, for example by optimizing the membrane surface area for metabolic exchange between the two cells. Once mitochondria became an intrinsic part of eukaryotic cells, their capacity for energy production could have conferred selective advantages on their host. However, the exact contribution of these organelles to the emergence of the complex features of eukaryotic cells remains unresolved.
Future efforts to elucidate the biological and physiological properties of Asgard archaea will be essential to increase our understanding of the emergence of eukaryotes. Although biochemical and structural studies of individual Asgard proteins, such as those by Akıl and Robinson, are likely to provide piecemeal insights, it is the ability to grow Asgard archaeal lineages in vitro that will ultimately unravel their obscure biology.
Nature 562, 352-353 (2018)
doi: 10.1038/d41586-018-06868-2
References
- 1.Williams, T. A., Foster, P. G., Cox, C. J. & Embley, T. M. Nature504, 231–236 (2013).
- 2.Raymann, K., Brochier-Armanet, C. & Gribaldo, S. Proc. Natl Acad. Sci. USA 112, 6670–6675 (2015).
- 3.Eme, L., Spang, A., Lombard, J., Stairs, C. W. & Ettema, T. J. G. Nature Rev. Microbiol. 15, 711–723 (2017).
- 4.Roger, A. J., Muñoz-Gómez, S. A. & Kamikawa, R. Curr. Biol. 27, R1177–R1192 (2017).
- 5.Spang, A. et al. Nature 521, 173–179 (2015).
- 6.Zaremba-Niedzwiedzka, K. et al. Nature 541, 353–358 (2017).
- 7.Akıl, C. & Robinson, R. C. Nature 562, 439–443 (2018).
- 8.Blanchoin, L., Boujemaa-Paterski, R., Sykes, C. & Plastino, J. Physiol. Rev. 94, 235–263 (2014).
- 9.Betts, H. C. et al. Nature Ecol. Evol. 2, 1556–1562 (2018).
- 10.Martin, W. F., Tielens, A. G. M., Mentel, M., Garg, S. G. & Gould, S. B. Microbiol. Mol. Biol. Rev. 81, e00008–17 (2017).
- 11.Grant, C. R., Wan, J. & Komeili, A. Annu. Rev. Cell Dev. Biol. https://doi.org/10.1146/annurev-cellbio-100616-060908 (2017).
Nenhum comentário:
Postar um comentário
Observação: somente um membro deste blog pode postar um comentário.